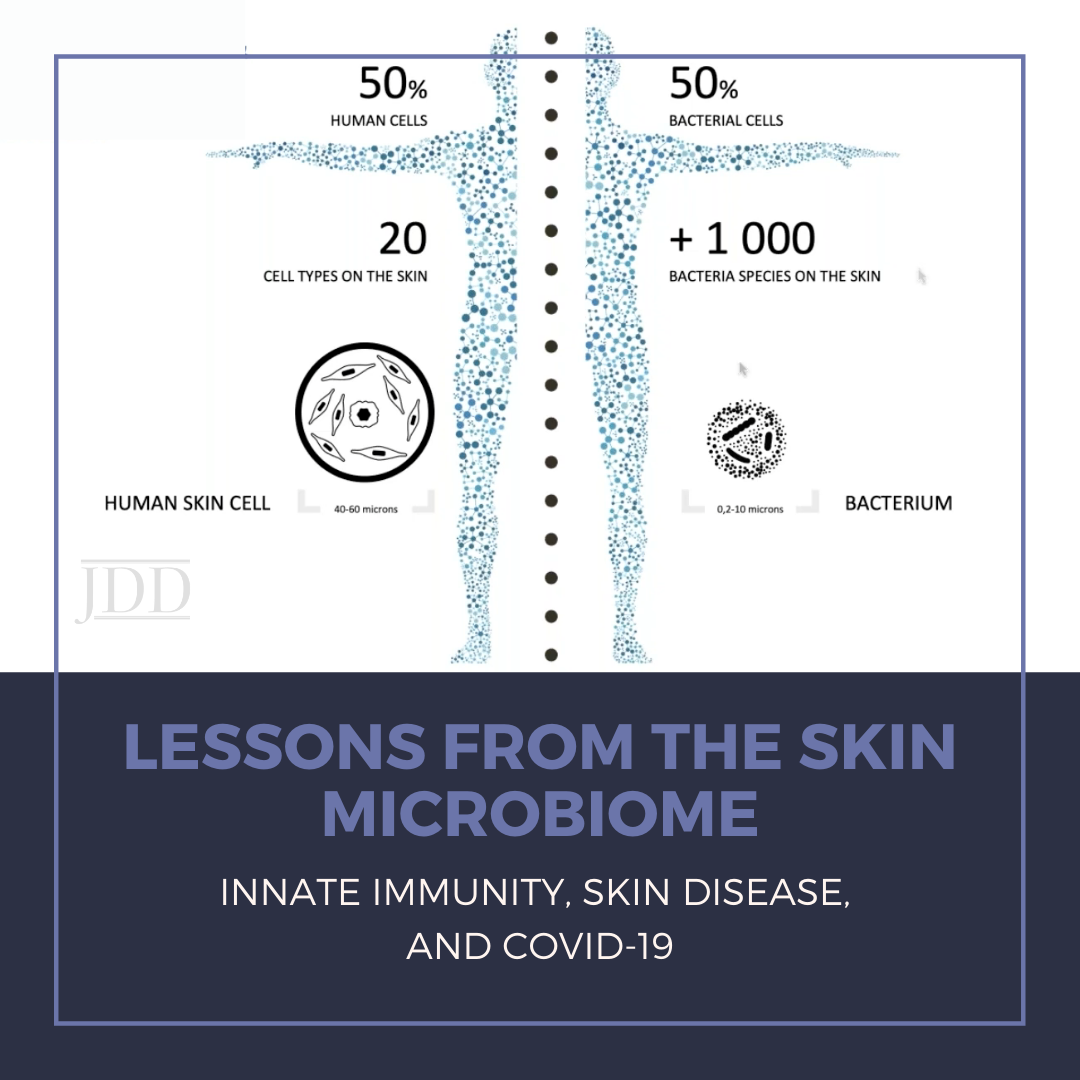
While I was beginning the process of “scrubbing in” to assist in the hemicolectomy case as an intern, I carefully squeezed the anti-septic chlorhexidine-soaked sponge and watched the brown solution drip its way down my forearms. I continued the ritual by cleaning under the fingernails and on every plane of the fingers, hands, and forearms multiple times. After all; “Clean Hands Save Lives,” as we have all heard over and over. This has especially been emphasized since the start of the COVID-19 pandemic, during which the Centers for Disease Control and Prevention (CDC) has told us that we should be diligently either washing our hands with soap and water or using hand sanitizer before eating or preparing food, before touching your face, after using the restroom, after leaving a public place, after blowing your nose, coughing, or sneezing, after handling your mask, after changing a diaper, after caring for someone sick, and after touching animals or pets.1 Yes, hand hygiene is certainly an important measure to prevent the spread of the deadly COVID-19 virus and to prevent us from coming into contact with potentially harmful microorganisms as well. But besides the physical barrier it provides, is our skin really otherwise pure and defenseless against pathogens? How could this be the case when, as humans, we are an ecosystem of interacting species, including bacteria and fungi, and only 50% of our cells are truly “human”?
This question was addressed by Dr. Richard Gallo, Irma Gigil Distinguished Professor and Chairman of the Department of Dermatology at the University of California San Diego, during his lecture “Lessons from the skin microbiome: innate immunity, skin disease, and COVID-19” presented as part of the George Washington University School Medicine and Health Sciences Dermatology Translational Lecture Series, hosted in partnership with the Journal of Drugs in Dermatology on December 17, 2020. Dr. Gallo noted how microbiome research to date has been largely limited in scope, disproportionately focusing on the gastrointestinal tract. However, taking into account all of the skin appendages, the surface area of the skin is larger, and the skin has a greater interface for microbial interactions than anywhere in the body. This is facilitated in part by the follicle, a structure that acts as a “microbial sink” by trapping and protecting the skin microbiome. Human skin has a natural antibiotic property, a defense system armed by many human anti-microbial peptides (AMPs). Twenty to thirty of these AMPs are functionally relevant, including cathelicidin (LL-37) and defensins. Certain AMPs may be constitutively expressed, while others are expressed for short periods of time, both serving to regulate the microbial community. Interestingly, bacteria also have AMPs, including bacteriocins. Understanding of bacterial AMPs has evolved over the years, with Dr. Gallo’s team contributing to the current theory that these AMPs are not just self-serving for the microbes themselves, but they are also expressed to protect us as humans. There is a complex system of AMPs in different epithelia and different “deployment systems” for AMPs designed on need. The skin, for example, has an excellent constitutive anti-gram-negative system and is also sometimes good against gram-positive bacteria. This is illustrated by an experiment conducted in which an individual touched S. aureus with a bare hand, waited 10 minutes, and then touched an agar plate which demonstrated bacterial growth on a replicate plating.1 The exact same experiment conducted with E. coli showed no growth on replicate plating.1 The control experiment, consisting of the same procedures with sterile surgical gloves, resulted in E. coli surviving on the surface of the glove, with S. aureus growing less after being transferred by way of the glove.1 These findings convey evidence suggesting that AMPs control surface microbial survival on human skin, and that we have more natural defenses than we previously thought! But what happens when this natural defense gets dysregulated?
The skin microbiome as it relates to atopic dermatitis (AD), a disease in which dysbiosis and barrier defects both play pivotal roles, was the next topic in Dr. Gallo’s discussion. S. aureus colonization is observed in most patients with AD and is correlated with disease severity. It is thought that S. aureus promotes disease through multiple mechanisms, including production of proteases and toxins which disrupt the epidermal barrier, allowing for S. aureus penetration into the skin. It is now understood that decreased cathelicidin and defensins, both suppressed by Th2 cytokines, and decreased bacteriocins expressed by bacteria, all act to effectively increase S. aureus colonization, even in non-lesional skin.2 When the microbiome becomes dysbiotic, S. aureusalong with specific strains of S. epidermidis become more prevalent, promoting inflammation, proteolysis, and Th2 skewing of the inflammatory response. In turn, these effects cause feedback to further inhibit AMPs made in the skin. This amplifying effect explains the chronic and cyclical nature of disease in AD. But can regularizing the microbiome improve barrier function and inflammation, thereby reducing Th2 skewing and correcting the cyclical nature of disease?
Dr. Gallo’s team has tried multiple methods of microbiome therapy. One example includes the approach of screening human skin for microbes that can inhibit S. aureus. Recently, with support from NIH/NAID, Dr. Gallo’s team conducted the first randomized, double-blind trial for microbiome therapy on the skin by taking an S. homidis strain (S.hA9) and applying it twice daily with Cetaphil cream on the skin of AD patients colonized with S. aureus. Patients treated with S.hA9 both on lesional skin and non-lesional skin showed a large decrease in their S. aureus colonization (unpublished data). Even when treatment was stopped, the S. aureus did not return (unpublished data). Microbiome sequencing of these individuals conveyed that, in patients treated with S.hA9, other species were proliferating on the skin relative to the S. aureus (unpublished data). Therefore, Dr. Gallo and his team concluded that S.hA9 not only inhibits S. aureus, but also permits other potentially beneficial species to survive on the skin of patients with AD. Furthermore, there was clinical improvement in a subset of these patients, as shown by statistically significant improvements in Eczema Area and Severity Index (EASI) and Scoring AD (SCORAD) metrics in patients who successfully had S. aureus killed (unpublished data). This study’s findings conveyed that bacteriotherapy with S.hA9 is safe, inhibits S. aureus, and that decreased skin inflammation in AD is correlated with killing of S. aureus. With all of our efforts to purify our skin and free it from pathogens, we may be learning the exact opposite approach can help us treat common skin disease such as AD!
What about rosacea? AMP abnormalities in rosacea include increased toll-like receptor 2 (TLR2), increased cathelicidin, increased kallikrein-related peptidase 5 (KLK5, a serine protease that activates cathelicidins), and altered processing of cathelicidin peptides. In rosacea, there is a defect in skin response to the microbiome and the environment, and receptors send an aberrant signal of host defense. This is illustrated by effects from sunlight, the number one reported trigger, as ultraviolet (UV) light can send out signals that are similar to those emitted by microbes. When UV light injures a cell, danger-associated molecular products (DAMPs) are released. Double-stranded DNA and RNA DAMPs may be bound by cathelicidin, which helps these DAMPs enter the cell.3 Furthermore, cathelicidin amplifies the inflammatory response of endothelia to UV products.3Cathelicidin works both by 1) direct receptor activation, and 2) facilitation of intracellular recognition. Simple amino acid substitution in the disrupted cathelicidin peptide causes loss of pro-inflammatory effect but does not affect bacterial killing, thereby dissociating its anti-microbial and anti-inflammatory activity.4,5 Gallo and colleagues used the proximity ligation assay to determine that active cathelicidin peptides would bind to the scavenger receptor SRB1 with the requirement of U1RNA, which joins to form a three-part molecular complex of sorts.5 He and his team concluded that specific structural requirements permit cathelicidin to enhance recognition of nucleic acids by scavenger receptors.5 Subsequently, nucleic acid is permitted entry into the cell, is recognized by TLR and cytosolic pathways, and activates inflammatory response.5 Under normal healthy conditions, U1RNA would not be able to activate inflammation, but cathelicidin performs “innate immune vetting”, in which its presence in the local environment determines whether or not host nucleic acids are pro-inflammatory in nature.5 Expression of cathelicidin is crucial in initiating appropriate inflammation in response to infection or injury, but also causes inappropriate inflammation in auto-inflammatory disease.5
Interestingly, understanding of dermatologic disease has also contributed to our understanding of other conditions involving immune dysfunction, the most relevant today of which is COVID-19. Other coronaviruses, such as Severe Acute Respiratory Syndrome associated coronavirus (also known as SARS, or SARS-CoV) and Middle East Respiratory Syndrome associated coronavirus (MERS), have similar life cycles to SARS-CoV-2, the culprit of the current COVID-19 pandemic devastating the world for the past year. Dr. Gallo worked closely with Dr. Gerard Wong, Professor in the Departments of Bioengineering, Chemistry, and the California NanoSystems Institute at UCLA, to investigate how SARS-CoV-2 induces the cytokine storm. They recently built a machine learning classifier to recognize sequences in the viral genome that mimic host AMPs. Using this machine learning program, they were able to screen the SARS-CoV-2 viral genome to see if there are any proteins made by these viruses, but not by other normal cold viruses, that more represented the structure of cathelicidin. Dr. Wong used structural models to show that some of the SARS virus peptides seemed to be able to assemble in these macromolecular structures with RNAs and DNAs, causing “innate immune vetting” and amplifying the immune response (unpublished data). They synthesized these innate immune peptides derived from machine learning and were able to show that Pep 6, 6s, and 11 show vetting activity on lung cells treated with a viral double-strand RNA mimic, poly I:C (unpublished data). In turn, they could predict peptide domains within SARS-CoV-2 that induced hyper-inflammation in lung cell lines as well as aortic endothelial cell lines (unpublished data). The evidence for SARS-2 peptides inducing innate immune vetting is an exciting byproduct of better understanding dermatology and pathophysiology of skin disease like rosacea and may lead to a better understanding of cytokine storm effects and innate immune dysfunction observed in COVID-19!
Understanding of the skin microbiome and host immune interactions, based on a culmination of growing literature in the field, is fundamental to understanding dermatology. Disease can result from not only too little immune defense leading to infection but also too much or altered defense leading to dysbiosis or aberrant signaling of host immune response. Understanding the ecological environment of our bodies is pivotal for practitioners treating patients, and we cannot underestimate the presence and significance of the inherent biochemical antimicrobial defenses in our skin. This exciting and expanding area of research has contributed not only to the understanding of dermatological conditions but also to other diseases involving dysregulation of immune responses, including COVID-19.
References:
-
- https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/prevention.html
- Gläser R, Harder J, Lange H, Bartels J, Christophers E, Schröder JM. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005 Jan;6(1):57-64. doi: 10.1038/ni1142. Epub 2004 Nov 28. PMID: 15568027.
- Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Science translational medicine. 2017 Feb 22;9(378).
- McCoy IV WH. “Shedding Light” on How Ultraviolet Radiation Triggers Rosacea. Journal of Investigative Dermatology. 2020 Mar 1;140(3):521-3.
- Braff MH, Mi‘i AH, Di Nardo A, Lopez-Garcia B, Howell MD, Wong C, Lin K, Streib JE, Dorschner R, Leung DY, Gallo RL. Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. The Journal of Immunology. 2005 Apr 1;174(7):4271-8.
- Takahashi T, Kulkarni NN, Lee EY, Zhang LJ, Wong GC, Gallo RL. Cathelicidin promotes inflammation by enabling binding of self-RNA to cell surface scavenger receptors. Scientific reports. 2018 Mar 5;8(1):1-3.
Source:
Lesson from the Skin Microbiome in Innate Immunity, Skin Disease and COVID-19. Presenter: Richard Gallo, MD, PhD is an Irma Gigli Distinguished Professor and the Chairman of the Department of Dermatology at the University of California San Diego. Hosted by the George Washington Universit y School of Medicine and Health Sciences in partnership with the Journal of Drugs in Dermatology. Watch the recording here.